
|
Manual
|
Peptide Properties Calculations:
This utility allows you to calculate the following properties of a peptide
based on the sequence:
- Calculate the estimated molecular weight of a polypeptide
- Estimate the partial specific volume of a protein and correct
the v-bar value to the temperature of your experiment
- Estimate the extinction coefficient at 280 nm of the denatured protein
according to the method by Gill and von Hippel.
- List the amino acid composition of a peptide
Note: This program will utilize the database backend of
UltraScan. This functionality allows you to save certain
files and data in a database, among them the peptide files analyzed
with this module. If the database system is not supported or available
on your system, you will get a warning message
when opening this module.
The peptide properties calculation module can be loaded in one of three ways:
- Click on "Calculate Protein MW and vbar"
in the Utility Menu from the Main Menu of UltraScan.
- Click on the vbar button from within any of the analysis windows in
UltraScan
- Through the database function "Commit
Data to DB:Buffer"
When using the second option, the temperature of the experiment will automatically
be used to correct the partial specific volume obtained from the sequence
analysis.
Explanation of fields and buttons:
|
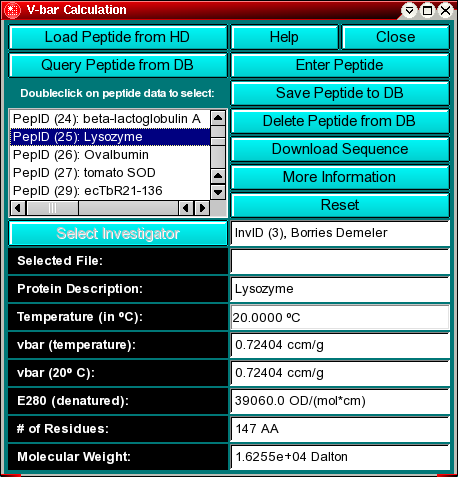
Peptide Sequence Loading:
You can load peptide sequences into this program by one of two methods:
- Load Sequence from HD
Use this button to load an ASCII sequence file from your harddrive. A file
dialog will open and allow you to search for *.pep files. The protein
file should have the extension ".pep" and should be located in your
UltraScan Root Directory.
- Query Peptide from DB
If you are using a database backend for UltraScan you can query
the database for peptide sequence files that have previously been committed
to the database. Sequence files available in the database will be listed in the listbox
once you click on this button. More information about various database functions is
available elsewhere.
|

|
Select Investigator: When searching for peptide sequences from
the database, you can limit your search to only those sequences belonging
to a specific investigator in the database. When you click on this button,
the investigator selection dialog
will open up and allow you to select the desired investigator. When the
sequence is loaded from the database, this option is disabled, since an
investigator is already associated with the selected sequence. Also,
when committing a sequence to the database an investigator needs to be
chosen to be associated with the sequence.
|
|
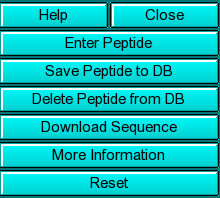
Help: Show this help file
Close: Close this function
Save Peptide to DB: Use this function to save a peptide sequence to
the database. Before storing a sequence in the database, you need to have
a copy of the sequence stored locally, for example, on your harddrive.
You can then select the sequence to be stored in the database from
a filedialog and after selection, commit the sequence to the database.
Delete Sequence from DB: Delete the selected sequence from the database.
This function requires administrator privileges.
Download Sequence: This button will open a Netscape window with a link to the Swiss Protein Database Search form. From
there, you can search and download the sequence for your protein (provided
it is already entered into the Swiss Protein Database). After finding
the appropriate sequence, save it as "text" with the extension ".pep"
in your UltraScan Root Directory
More Information: Clicking on this button allows you to
review the amino acid composition and the vbar calculation results. The results
are always written to a file called <peptide file
name>.res
Peptide Reset: Clear the loaded peptide sequence from memory
|
|
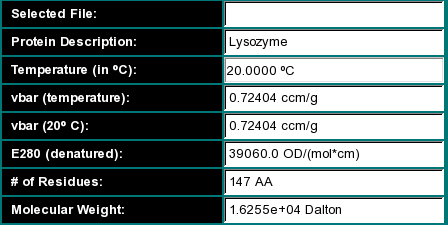
Selected File: The location of a Swiss Protein database sequence entry
file that can be imported directly into UltraScan. See
also the information on the "Download Sequence" button. This field is
blank if the sequence was retrieved from the database, and only applies
to sequences loaded from the harddrive.
Protein Description: The name of the protein
Temperature (in oC): The temperature of the
experiment
vbar (temperature): The partial specific volume of the
protein corrected to the experimental temperature
vbar (20 oC): The partial specific volume of
the protein at 20 oC. All vbar calculations are based on the method
by Cohn and Edsall (Proteins, Amino Acids and Peptides as Ions and Dipolar
Ions. New York. Reinhold, 1943.)
E280 (denatured): The extinction coefficient at 280 nm
for the peptide in a denatured form based on the method by Gill and von Hippel
in Anal. Biochem. 182:319-326 (1989). This number is based on the 280
absorbance of Tyrosine, Cysteine and Tryptophan in 6 M urea. Use at your own
risk - measuring this value will be more accurate. UltraScan
will require extinction coefficients for the calculation of equilibrium
constants.
# of Residues: The number of amino acids in the sequence
Molecular weight: The theoretical molecular weight obtained
by summing the partial amino acid molecular weights, and subtracting one less
water molecules than amino acid residues present.
|
You can generate your own peptide sequence file, and import them into
UltraScan, provided the sequence file follows the correct
sequence file format:
Required Fields in Database file: When generating your own sequence
file, you will have to follow the Swiss Protein Database file format convention.
However, not all fields are required for correct sequence parsing. The fields
that are required are:
- "DE" - Sequence Description tag. Prepend any description line with
this tag to identify the description of the sequence. Note: Only the first
line is parsed, all others are ignored. There can only be one sequence/file.
- "SQ" - Sequence Entry. Use this tag to identify the start of
the sequence in a line preceeding the actual sequence, In the line following
the sequence place the end-of-sequence marker "//"
Sample Sequence file with minimum entry fields:
DE LYSOZYME C PRECURSOR (EC 3.2.1.17) (1,4-BETA-N-ACETYLMURAMIDASE C)
SQ SEQUENCE 147 AA; 16238 MW; 81E85743FF579468 CRC64;
MRSLLILVLC FLPLAALGKV FGRCELAAAM KRHGLDNYRG YSLGNWVCAA KFESNFNTQA
TNRNTDGSTD YGILQINSRW WCNDGRTPGS RNLCNIPCSA LLSSDITASV NCAKKIVSDG
NGMNAWVAWR NRCKGTDVQA WIRGCRL
//
www contact: Borries Demeler
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
http://www.ultrascan.uthscsa.edu
Last modified on January 12, 2003.






