
 |
Manual |
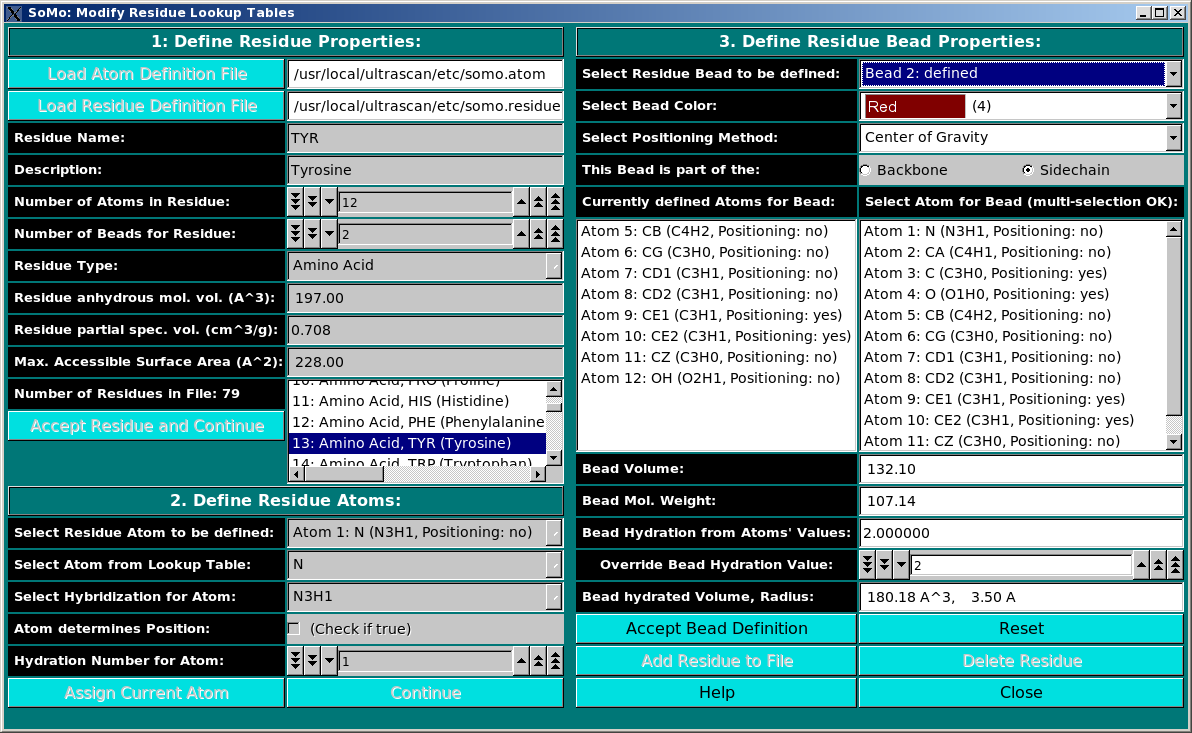
This module is used to define all residues that can be found in PDB files. In this module, you can also define the rules which are used to convert them into beads. You can add new residues or modify the properties of the existing ones. Three panels are available:
Panel 1: Define Residue Properties:
 |
|
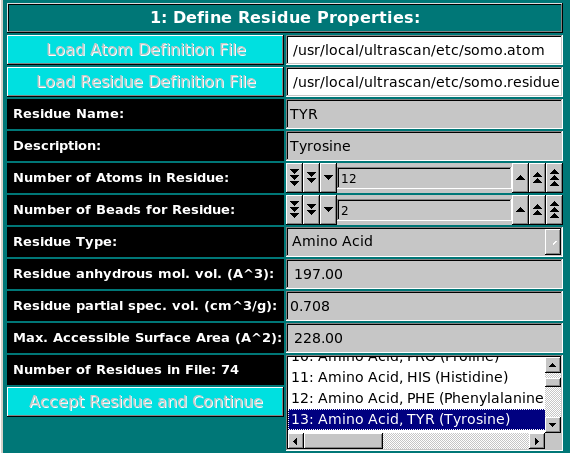
In this panel, you can add or modify the general properties of a residue.
To add a new residue, you must first load the atom table
containing the atomic groups, as defined in the PDB file (N, CA, CB, etc.),
that make up the residue.
The Residue anhydrous mol. vol. (A^3) field has to be filled with a
value usually derived from crystallography (see Tsai et al., J. Mol. Biol.
290:253-266, 1999; Nadassy et al., Nucl. Acid Res. 29:3362-3376, 2001; Voss
and Gerstein, J. Mol. Biol. 346:477-492, 2005; Perkins, Eur. J. Biochem.
157:169-180, 1986). This is the volume that defines the total anhydrous volume
of the bead(s) that will be used to model the residue, and for non tabulated
entries it can be computed using on-line programs such as the 3V Contact Volume Calculator (http://www.molmovdb.org/cgi-bin/3v.cgi; a 0
Angstrom probe radius should be used). For ions, we used the values in Table I of
Kiriukhin and Collins (Biophys. Chem. 99:155-168, 2002).
When all fields have been filled, the residue is accepted by pressing the Accept Residue and Continue button, and the Number of Residues in File field is consequently updated. |
Panel 2: Define Residue Atoms:
 |
|
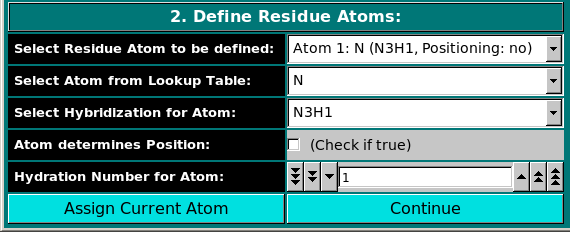
In this panel, the atoms making up the residue are chosen, using the
atom table previously selected. The atoms are numbered
sequentially up to the value entered in the Number of Atoms in Residue
field in the previous panel, and can be chosen from the Select Residue Atom to be defined
pull-down menu.
|
Panel 3: Define Residue Bead Properties:
 |
|
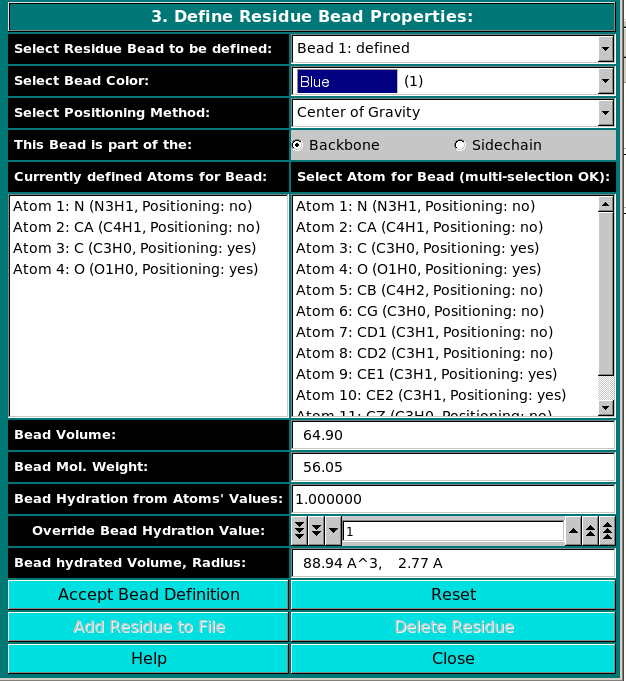
In the last panel of this module, the atoms in each residue are assigned
to bead(s), and each bead's properties are then defined.
Center of gravity: between the atoms assigned to the bead and marked
"yes" in the Atom determines Position checkbox in Panel 2;
Each bead needs then to be defined as belonging to the backbone or to the sidechain of the molecule by clicking the appropriate box in the This Bead is part of the: field. This determines at which sequential stage the bead will be processed during overlap removal in the SoMo method. The next step is the assignment of atoms to the bead. In the Select Atom for Bead (multi-selection OK) window, you can see the list of all the atoms that belong to the residue, as defined previously in Panel 2. By clicking on an atom, it will be also displayed in the Currently defined Atoms for Bead window to the left. Once all atoms belonging to a bead have been selected, the bead is accepted by clicking on the Accept Bead Definition button. The Bead Volume field defines the anhydrous volume (in A3) assigned to the bead. IMPORTANT: the sum of the volumes of the bead(s) defining a residue MUST match the Residue anhydrous mol. vol. entered in Panel 1. The program will NOT let you save the residue/bead definition if there is a discrepancy between these two values. The values present in the current definitions of the amino acid residues were derived from the crystallographic analysis (see above for references). When a molecule needs to be subdived in more than one bead, the volumes of the various parts can be determined using the 3V Contact Volume Calculator (see above). However, the parts' volumes will NOT add up to the entire molecule volume, and MUST be then proportionally rescaled. The volume of all other entries in the current somo.residue table were determined in this way. The next field, Bead Mol. Weight is for visualization only, and will display the bead molecular weight, calculated from the values (taken from the atom table. Likewise, the subsequent field, Bead Hydration from Atoms' Values, will show the theoretical number of water of hydration molecules assigned to the bead, based on the sum of the atoms' values entered in Panel 2. However, since the atomic values are likely not defined for most residues, a global value for each bead can be entered in the next field, Override Bead Hydration Value. Most beads' values currently present in the somo.residue file were derived from literature. For carbohydrates, we used the values of Shiio (J. Am. Chem. Soc. 80:70-73, 1958). For ions, we used the values in Table I of Kiriukhin and Collins (Biophys. Chem. 99:155-168, 2002). For the other residues, and for further information, see Brookes et al., Eur. Biophys. J. in press, 2009, and Rai et al., Structure 13:723-734, 2005. The volume of these water molecule(s) (defined under the Miscellaneous Options; see here) will be later automatically added to the anhydrous volume of the bead to define its hydrated volume and radius. Finally, in the last field, Bead hydrated Volume, Radius, is again for visualization only, and will display the bead's hydrated molecular volume and hydrated radius (derived by summing to the anhydrous Bead Volume the volume of the water of hydration molecules, see above). If the residue is represented by more than one bead, the process is repeated until all beads have been processed. The residue can then be added to the file by clicking on the Add Residue to File button. The process can be interrupted at any stage by pressing the Reset button. To Edit an already defined residue, double-click on its name on the list
(that can be scrolled) present in the window next to the Number of residues in
File field in Panel 1. All the stored properties will then appear
first in the Panel 1 fields, which can then be updated. By clicking on the
Accept Residue and Continue button, the properties listed in Panel 2
become then accessible, and can be likewise edited. Finally, the Panel 3
options will become again available for editing by clicking on the
Continue button in Panel 2. Accept Bead Definition
and Add Residue to File buttoms will then allow to update the
residue table (somo.residue).
|
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The
latest version of this document can always be found at:
http://www.ultrascan.uthscsa.edu
Last modified on February 1, 2010.