
 |
Manual |
The Monte Carlo Simulation program allows you to generate the equivalent of multiple experimental observations by simulating experimental noise using a random number generator. Each simulated observation can then be refitted and provide a new parameter vector of estimated best fit solutions. The collection of parameters can then be used to derive statistical measurements that allow you to assign a confidence to the obtained parameter value.
Explanation of Fields:

With this button, you can select an output file for the Monte Carlo iterations. By default, the name of the run (velocity) or the project name (equilibrium) is used and ".mc" are appended. In the case of velocity experiments, the cell and wavelength number are also included in the output filename, to avoid overwriting outputs from different cells of the same run. For equilibrium experiments, the model number of the equilibrium fit is also incorporated into the filename.
You can also enter a filename by hand in the dialog field. Make sure to enter the full path, and not just the filename. By clicking on the checkboxes for the append or overwrite mode, you can select if the output file is created new (overwrite) or if information is simply appended to the information that has been previously collected (either in a previous Monte Carlo session or in a simultanous Beowulf session).
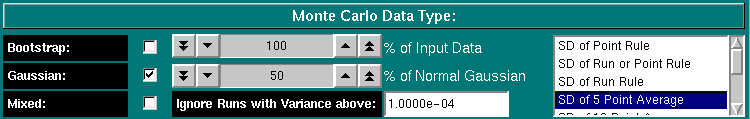
This function allows you to determine the composition of your simulated data. Your options are "Bootstrap", "Normal Gaussian" or "Mixed". In the normal Gaussian mode (the default), synthetic data will be generated based on the selected data generation rule. The following options exist:
The bootstrap method utilizes a randomly distributed selection of residuals from the original fit, which will be added onto the best fit values. These randomly selected residuals can be mixed with a user-selected percentage of the residuals from the original fit. Use the "% of Input Data" counter to set the percentage of "bootstrapped" residuals vs. original residuals included in the bootstrapped Monte Carlo. A setting of 0% would be tantamount to fitting the original data, while a setting of 100% would randomly pick and rearrange the residuals from the original fit to create a dataset that has the same residuals, but all are placed at different points in the data.
If you select "Mixed", you can specify the percentage of points that will have a random normal Gaussian noise distribution added, the rest of the points will have bootstrapped residuals. For example, if you select 75% for the setting "% of Normal Gaussian", then 75% of the datapoints will have random Gaussian-distributed residuals that are generated based on the standard deviation of the original fit, the rest of the datapoints will be bootstrapped according to the setting under "Bootstrap" and "% of Input Data". If the percentage of Gaussian generated points is larger than 0%, the selected percentage of Gaussian datapoints will be generated with the same Rule set as selected in the pure Gaussian method.
Note:
All percentages listed will be approximate only, since a random number
generator cutoff scheme is used to allocate percentages. Please consult
the Monte Carlo Tutorial for
suggestions and hints on which method and rule is the most appropriate
for a given application.
On occasion, a fit may fail to converge, in which case the variance of this Monte Carlo iteration will be unreasonably large. Setting a cutoff value here allows filtering of those errant iterations. The value can be adjusted after all iterations have completed or during the run. Clicking on the "Update Parameter" button (see below) will apply the new limits.

Sometimes, when error surfaces are ill-conditioned, a fitting session may become stuck in a local minimum, preventing the proper survey of the entire error surface. In such cases it often helps to add a little random noise to the original parameter guesses to start the fit at a different position in each iteration. This way chances for finding a global minimum are improved. The disadvantage of this method, however, is that more iterations will be required to converge the fit. The are three choices for how to apply the initial guesses:
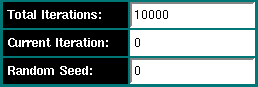
Under the field "Total Iterations" you can enter a fixed number of total iterations that need to be completed before the Monte Carlo session is finished. I recommend at least 10,000 iterations to obtain reliable results. The field entitled "Current Iteration" will be updated with each iteration performed by this session. If other sessions (SMP or Beowulf) are writing to the same output file, this information will be captured automatically and the "Current Iteration" field will be updated automatically. The "Random Seed" field will list a computer-generated seed (the process ID XORed with the system time) to generate a somewhat random seed for the randomization of the Monte Carlo data type. You can also enter an arbitrary integer value if you prefer to use your own seed. Random seeds are necessary since they prevent using the same random sequence start point in different Monte Carlo sessions. To make sure that no parallel session utilized the same seed, you can compare the entries in the second column of the output file, they contain the random seed used in the Monte Carlo session.

This listbox can be used to select the Monte Carlo statistics for a specific parameter that is being fitted in the Monte Carlo analysis. Double - click on the desired parameter to view the parameter distribution as well as other statistics.

Each time you select a different parameter with the listbox above, the Mean Value (and all other statistics) are recalculated. The Mean Value is displayed in the top line. In the next line, you can enter the number of bins used in generating the frequency distribution of the parameter values that have been generated in the Monte Carlo analysis. The smaller the bin number, the larger the bin size and the more datapoints will be counted in each bin. The last line lists the number of datapoints available from previous iterations that have already been completed.

"Update Parameter": The Monte Carlo output file is read and the current parameter is updated. This function is useful if you use the software in a Beowulf or SMP system with multiple sessions writing to the same file. In that case you can monitor the progress of the other sessions from the local session.
"Statistics": This button will call the Monte Carlo Statistics window for an update of the statistics for the currently selected parameter. With each iteration performed by the local session, this window is updated automatically if it remain opened. Clicking on the "Update Parameter" button also will update the statistics window. No statistics are calculated until at least 100 iterations have been completed.
"Print Histogram": Print a copy of the current histogram to your printer.
"Save Data": Save a copy of the statistics and histogram information to an ASCII file. See File Structures and Formats for details on the file format used.
"Start" and "Stop": These buttons are used to control the Monte Carlo Fitting process. You can start or stop the process. When stopped, the current iteration will complete before the fitting is altogether stopped. The buttons are logically linked, and disabled when not appropriate for use.
"Help": This help menu.
"Close": Close the application.

This plot shows a histogram of the distribution of all parameter values, sorted into bins of size (maximum value - minimum value) / (number of bins). The number of bins (and hence the size of bins) can be adjusted with the Monte Carlo information field "Number of Bins:" (see above). No plots or statistics are shown until at least 20 iterations have been completed.
On occasion a fit may fail to converge and will produce unreasonable answers, causing a distortion of the histogram plot. In such a case you can graphically crop the subset of data with reasonable values by dragging the mouse over the desired region in the plot. Only the x-axis limits of the bounding rectangle will apply. You can repeat this cropping process repeatedly, until all unreasonable data have been eliminated. In most cases unreasonable values (for example, of the variance) translate into unreasonable values in the histograms of other parameters. As you crop datapoints in the histogram of one parameter, the corresponding datapoints from all other parameters for the run that gave unreasonable values will be eliminated as well. The adjustment of iterations will be reflected in the field for "# of Data Points" and "Current Iteration".
If you make a mistake, you can reset the distribution by clicking on "Update Parameter", which will re-initialize the parameter distributions from the file.
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
Last modified on January 12, 2003